Dienstag, 20. Dezember 2011
Most useless installation instructions
So this guy wants to install phpBB. From phpBB: "If you see a "Could not connect to the database" error, this means that you didn't enter the database data correctly and it is not possible for phpBB to connect. Make sure that everything you entered is in order and try again. Again, if you are unsure about your database settings, please contact your host." Oh really? No, I enter wrong stuff ON PURPOSE because I enjoy spending time reading error messages YOU MORONS!
Freitag, 11. November 2011
Talks: Comments
Seminar at a group where nobody knows your work:
- Most important point: the talk is not for you, it's for the audience. The talk has to be designed such that the audience enjoys it, not you
- Who is the audience? What do they know? How much introduction is required such that it becomes understandable what I'm saying?
- Once I know who the audience is going to be: draft the talk in four slides. Condense the material
- Readable graphs (large units/labels, thickness of lines), i.e. if graphs are not prepared nicely, it shows that the preparation of the talk was not a priority
- Better the audience understands a little a bit, instead of it totally does not understand anything
- Simple Rule of thumb 1: Not more than two messages per slide
- Simple Rule of thumb 2:
Not more than six bullet points, not more than 6 white-spaced expressions per bullet pointNo bullet points, substitute bullet points by figures Dont read the bullet points, memorize them. Go through them while facing the people, not the slidesAddress each figure and make it clear why it is there- The slides dont substitute the talking. The talking is the main message carrier, the slides should allow to assist when explaining. Thats why text is not required, instead figures
- Dont base the talking on the slides, base the slides on the talking.
- Timing (excluding questions): 5% personal introduction: "who is this guy?", 10% content introduction, 70% content, 10% summary/conclusions, 5% aknowledgements
- Start practicing the talk at least
3 daysa week ahead. - Avoid text
- In the start of the talk, it's fine to show something unrelated so people remember you
Dienstag, 8. November 2011
Donnerstag, 3. November 2011
GNUPlot 002: EPS Bounding Box
What I now do is to use gnuplot terminal "postscript enhanced" and print to ".ps" file. Then I use the tool ps2eps, which generates a better bounding box. The call is
ps2eps -f -B -l -R + data.ps
-f: forces overwrite of existing eps files, -B recalculates bounding box, -l adds a bit to it, so it does not terminate on the labels, -R + sometimes rotation is required. No output file is required.
Then, the .eps file is opened in preview and saved to PDF. If using "convert" again PDF will have a too large white space margin, probably there are some options to adjust this, havent found them yet.
Another option (Credits: Janus)
http://www.gnuplot.info/scripts/files/fixbb
Mittwoch, 2. November 2011
Latex 001: Making a table cover the page width
\begin{table*}[htbp]
\begin{tabular*}{\textwidth}{l@{\extracolsep\fill}lrrrr}
\\\hline
1B2V & 32 & -4.06 & 4.23 & 4.72 & 7.30\\
1DUK & 64 & -0.35 & 3.94 & 4.45 & 4.80\\
1DEK & 137 & 1.88 & 4.87 & 5.19 & 5.80\\
\end{tabular*}
\caption{Some values.}
\label{tab:values}
\end{table*}
Dienstag, 1. November 2011
CGI 001: Combining Javascript and CGI
<html>
<script type="text/javascript">
function helloworld()
{
document.write("Hello World.");
}
</script>
<body>
<input type="button" value="Hello World." onClick="helloworld()">
</body>
</html>
</pre>
Including CGI:
<html>
<script type="text/javascript">
function helloworld()
{
return true;
}
</script>
<body>
<form action="./hello.cgi" onSubmit="return helloworld()">
<input type="submit" value="Hello World.">
</form>
</body>
</html>
The CGI part in Python#!/usr/bin/python print "Content-type: text/plain\n\n" print "Hello World."The Javascript function is nothing but a pseudo-function. It does nothing, but control of flow does go through it, otherwise the form is not submitted to the CGI.
Important to note is that the CGI script access the element name property, not the ID.
Mittwoch, 14. September 2011
GNUPlot 001: Postscript postproduction
Settings were:
set terminal postscript
set output 'outfile.ps'
set size 0.6,0.6
set xlabel "Reaction Coordinate"
set ylabel "dE[kcal/mol]"
plot 'data.dat' with lines lw 4
set output
If i open this file in preview, the file is oriented 90° counter-clock wise. This can be fixed with 'ps2eps'.
The command I used was:
ps2eps -f -R + -B -l data.eps
Samstag, 3. September 2011
Python 003: Summing specific items of nested lists
An example might be
a=[[2,4,6],
[4,2,5],
[2,5,6]]
where we're interested in the sum of 6+5+6. In Python this can conveniently be done using the reduce/lambda pair of functions:
>>> reduce(lambda x,y: x+y, [i[-1] for i in a])
17
>>>
Samstag, 30. Juli 2011
Schey 003: Surface Integral over Half Sphere
$\left[ \begin {array}{ccc} \sin \left( \phi \right) \cos \left(
\theta \right) &-R\sin \left( \phi \right) \sin \left( \theta \right)
&R\cos \left( \phi \right) \cos \left( \theta \right)
\\ \noalign{\medskip}\sin \left( \phi \right) \sin \left( \theta
\right) &R\sin \left( \phi \right) \cos \left( \theta \right) &R\cos
\left( \phi \right) \sin \left( \theta \right) \\ \noalign{\medskip}
\cos \left( \phi \right) &0&-R\sin \left( \phi \right) \end {array}
\right]$
Dienstag, 12. Juli 2011
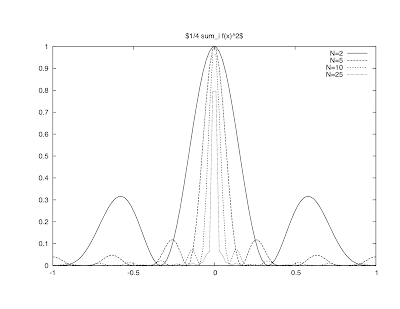
Web 002: Interactive Figure
Montag, 11. Juli 2011
Web 001: Setting up Apache and Running phpinfo() on Mac OS X
- cp /etc/apache/http.conf /etc/apache/back_http.conf # No backup, no mercy.
- sudo vi /etc/apache/http.conf
- You need to change two lines: Locate "DocumentRoot" and set the path behind it to '/Users/yourUserHomeDirectory/Sites'. Then locate the <Directory "/Library/WebServer/Documents"> line and change the path to the one you entered above.
- Remove the '#' sign infront of the line starting with "LoadModule php5_module"
- Save the file and exit.
<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN"Then enter the following into a file called phpinfo.php and place it in the ~/Sites directory as well.
"http://www.w3.org/TR/xhtml1/D
TD/xhtml1-transitional.dtd">
<html>
<head>
<title>Index Testing PHP</title>
</head>
<body>
<br /><br />
/Users/yourUserHomeDirectory/Sites/index.html
<form method="POST" action="phpinfo.php">
<input type="submit">
</form>
</body>
</html>
<?php phpinfo(); ?>
Freitag, 8. Juli 2011
Schey 002: Surface Integral
> 2*Pi*a^2*int(r*(a^2-r^2)^(-1/2),r=0..a) assuming a>0;
3
2 Pi a
>
Schey, Ex. II.5,b.
Donnerstag, 7. Juli 2011
Shell 007: Zero Padding
This can be done by using
for i in 000{1..9) 10
do
echo $i
done
Freitag, 24. Juni 2011
Jmol 003: Arranging Protein on NW Surface
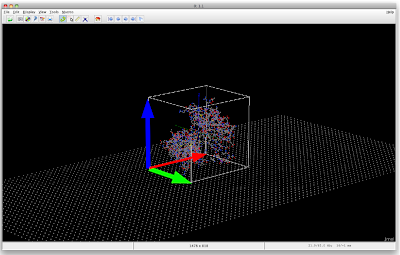
load 1PDB.pdb
load APPEND nwSurface.xyz
model all
axes on
select protein
boundbox {1.1} on

draw xVec vector {-58.19/2.0, -47.57/2.0, -54.71/2.0} {58.19, 0.0, 0.0} width 5.0 color red
What I now require is to have the vectors rotate together with the protein, so maybe the easiest is to have the vectors be part of the protein structure.
Mittwoch, 22. Juni 2011
Jmol 002: Control over Two Structures
<!DOCTYPE HTML PUBLIC "-//W3C//DTD HTML 4.01 Transitional//EN"This results in the figure below.
"http://www.w3.org/TR/html4/loose.dtd">
<html>
<head>
<title>UI Controls example</title>
<script src="./jmolDir/Jmol.js" type="text/javascript"></script>
</head>
<body>
<script type="text/javascript">
jmolInitialize("./jmolDir");
jmolApplet(400, "load 1AVD.pdb;\
load APPEND nwSurface.xyz;\
axes on;\
boundbox on;");
</script><br />
</body>
</html>
 A protein structure is placed above an array of carbon atoms which are supposed to illustrate the surface of a nanowire. What I would like to do now is to be able to rotate the protein with respect to the surface (i.e. keeping the surface in the same orientation regarding the indicated coordinate system).
A protein structure is placed above an array of carbon atoms which are supposed to illustrate the surface of a nanowire. What I would like to do now is to be able to rotate the protein with respect to the surface (i.e. keeping the surface in the same orientation regarding the indicated coordinate system).Unfortunately, I dont know how to include Jmol on Blogspot, otherwise I naturally would have included it directly here as well.
The input from Bob Hanson at the Jmol Mailing list is actually pointing me into exactly the correct direction. This script was tested in a local version of Jmol but it should just as well be working in a browser.
load 1PDB.pdb
load APPEND nwSurface.xyz
select protein
set allowRotateSelected True
set dragSelected True
Now using <ALT>+<Left_Mouse> the user can rotate the protein structure without changing the surface and <ALT>+<SHIFT>+<Left_Mouse> to drag the structure over the surface.
Dienstag, 21. Juni 2011
Jmol 001: Loading a Structure File from Disk Through CGI
#!/usr/bin/pythonNote, all HTML code had to be escaped in order to be posted on the blog.
print "Content-type: text/html\n\n"
import cgi
import cgitb
cgitb.enable()
def getJmolPage(target):
newpage = ''
newpage += '<html>\n'
newpage += ' <head>\n'
newpage += ' <title>Load PDB Form</title>\n'
newpage += ' <script src="./jmolDir/Jmol.js" type="text/javascript"></scri
pt>\n'
newpage += ' </head> \n'
newpage += ' <body> \n'
newpage += ' <script type="text/javascript">\n'
newpage += ' jmolInitialize("./jmolDir");\n'
newpage += ' jmolApplet(400, "load ../x_output/%s.pdb");\n'
newpage += ' </script><br />\n'
newpage += ' </body>\n'
newpage += '</html>\n'
return newpage % (target)
if __name__ == '__main__':
form = cgi.FieldStorage()
target = form['PDBID'].value
print getJmolPage(target)
Donnerstag, 16. Juni 2011
Reading
File Permissions: Good short summary of dangers in web programming
HTML/JavaScript: Displaying text when clicking on checkbox.
Science
Ising Model: Derivative of Magnetization, Source, Phase transition
Lipases: Chemical Industry Review.
Dienstag, 14. Juni 2011
PyMOL 004: Illustrating a periodic system
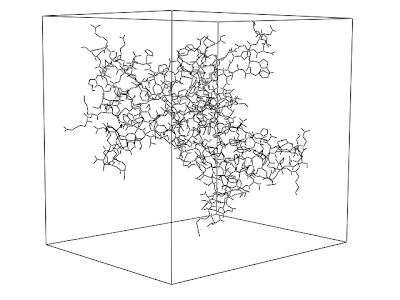
supercell 40,4,1, withmates=1at the PyMOL prompt.
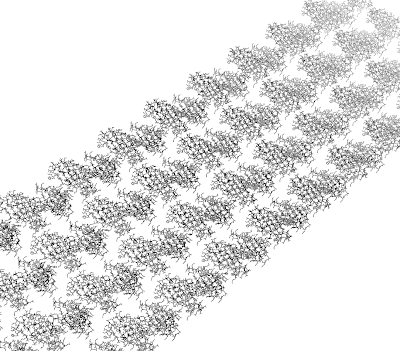
After aligning all proteins on the surface, the output is the following.

Montag, 30. Mai 2011
Shell 006: Replacing String with incrementing index
header
foo
foo
foo
header
bar
bar
bar
header
baz
baz
baz
where we want the "header" be replaced by "model {1,2,3}".
I was pointed out to a couple of solutions over at LinuxQuestions.org
An awk solution.
$ awk 'BEGIN { cntr = 0 } /header/ { cntr++ ; print "model", cntr } !/header/ { print $0 }' infile
where 'infile' contains the data.
A bash solution.
#!/bin/bash
filename=data
index=1
for entry in `grep -n model $filename`
do
line=`echo $entry | awk -F":" '{print$1}'`
sed -e "$line s/model/model $index/" -i $filename
index=$(($index + 1))
done
An even shorter awk solution.
awk '$0 = /header/?"model "++c:$0' file
A one line bash solution
unset count; while read line; do [[ $line =~ header ]] && line="model $((++count))"; echo "$line" >> oufile; done < infile
Great to have the support from the online community.
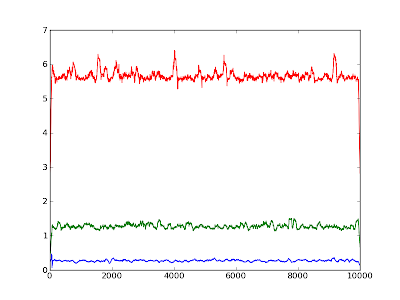
Python 002: Averaging over values
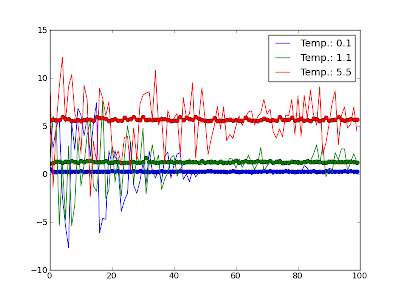
Eav = array([reduce(lambda x,y: x+y, i)/len(i) for i in [E[i:i+segm] for i in range(0, len(E), segm)]] )
I have been pointed out to a solution making use of the numpy.convolve method.
means = convolve(E, ones(100)/100.0, 'same')
Preparing a Presentation
- Who will be my audience? This is what should guide in the design and layout of the talk. It decides over what degree of detail to present, ranging from raw data and formulas to graphs of the basic relationships over to cartoon representations of the physical problem.
- How many messages does one slide carry? It should not be more than two. Usually, things are complicated. It is preferred to broadcast fewer information, but have that information understood instead of broadcasting masses and nothing is understood: "If they understand a little, that's better than if they didn't understand a lot." If one slide refers back to a previous one, include it on the slide in a small version to help keeping it in memory. It is also a good idea to keep a couple of extra slides which contain more details, just in case.
Sonntag, 29. Mai 2011
Griffith, Ex. 2.7
The solution (without inserting values) is computed with Maple.
The question would be now, how to get the integral from x=-1 to x=1. I need to tell Maple that z > R. How can I do that?
One a side note, it seems as if I can not view the MathML in Safari. Why is that?
Montag, 2. Mai 2011
Shell 006: Nested Calls
Mittwoch, 13. April 2011
Shell 005: String processing
~/shell $ ls -l
00_01_boxpdb.py
00_01_realign.py
00_02_callPDB2PQR.py
00_03_getPointCharges.py
00_04_getCharge.py
01_01_connect.py
How can the leading terms be removed?
~/shell $ for i in 0*
> do
> mv $i ${i:6}
> done
The syntax is ${i:FROM:BY}. The indexing starts from 0. Also
# Is it possible to index from the right end of the string?
echo ${stringZ:-4} # abcABC123ABCabc
# Defaults to full string, as in ${parameter:-default}.
# However . . .
echo ${stringZ:(-4)} # Cabc
echo ${stringZ: -4} # Cabc
# Now, it works.
# Parentheses or added space "escape" the position parameter.
Shell-004: Selective Listing
ls -l|grep ^d|awk '{print $8}'
Grep matches the beginning of the line output by 'ls' with 'd', which indicates a directory. " 'awk {print $8}' " then prints the 8th column of the output (so the directory name is available again).
Dienstag, 12. April 2011
Vim 005: Efficient formatting
export PYTHONPATH=$HOME/shellscripts:$HOME/tree/lib64/python2.4/site-packages:$HOME/kemibin/2010/woche_03/wave/
and it is difficult to read. It would be nice to have all directories added to the path on separate lines. In Vim, this can be done by issueing:
:s/:\$/:\rexport PYTHONPATH=\$PYTHONPATH:\$/g
\r is the line feed character (apparently it works on Linux, even if its the Windows line feed character). The common Unix linefeed character is \n. \$ escapes the dollar sign so it can be found and replaced/inserted (it would be considered the end of the line if not escaped). Calling the command returns
export PYTHONPATH=$HOME/shellscripts:
export PYTHONPATH=$PYTHONPATH:$HOME/tree/lib64/python2.4/site-packages:
export PYTHONPATH=$PYTHONPATH:$HOME/kemibin/2010/woche_03/wave/
There remains a colon on all but the last lines. Replacing this would require the command to recognize if it is operating on a line which is not the last one of the selection.
Vim 004: Pasting into Vim
In Vim, use
:set paste
to enable pasting from the clipboard.
Vim 003: Deleting all Lines of given Kind
:d/IDENT/g
Sonntag, 10. April 2011
Shell 005: Redirect Part of a File
for i in file-*.log
do
sed -n '0,/DONE/!p' $i > ${i/file/new}
done
"!" tells it to ignore everything from the start of the file "0" to the first line containing the keyword "/DONE/", then p makes it print the rest, with a shell redirection of the output to a new file.
Regards to David the H. at LinuxQuestions for pointing this out to me.
Freitag, 8. April 2011
Vim 002: Submitting Vim Command from the shell
vi "+:%s/+0/+1/g" "+wq" $i
Notice the '+' operator within the command string. Also notice the 'wq' command to save and "exit" Vim once it finished its job. This is an excellent way of processing a batch of files.
Shell 003: Tail
count=`cat file.dat|wc -l`
tail -n -$((count-40)) file.dat > newfile.dat
Notice the backtics to evaluate the cat and wc calls in the first line. Also notice the '-' sign in front of the numeric argument to the tail command.
Dienstag, 5. April 2011
Montag, 4. April 2011
Shell 004: Arithmetic Extension
mzh @ ~/shell $ for i in {1,2,3}
> do
> tmp=$((i*i))
> echo $tmp
> done
1
4
9
I found it here.
PyMOL 003: Displaying Atom Coordinates
PyMOL>iterate_state 1, sele, print name, x, y, z
Sonntag, 3. April 2011
GAMESS 002: Copying to a new job type
mzh @ ~/shell $ for i in a-{1,2,3}-opt.log
do
OPTLOG=$i
HESLOG=${OPTLOG/opt/hes} # Rename jobtype
HESINP=${HESLOG/log/inp} # Rename extension
grep -i -A 29 "equilibrium" $OPTLOG|tail -n 26 >> $HESINP
done
Now a more tricky part, transfering the Hessian matrix from the GAMESS output .dat file to the SADPOINT job file. How can this be automated in a smart way?
This line of code is the solution:
mzh ~/shell $ awk '/ \$HESS/,/^ \$END/' hess.dat >> sad.inp
I dont know what the ',' does. Note the quotes.
mzh ~/shell $ ruby -ne 'print if /\$HESS/../\$END/' file
or
mzh ~/shell $ f=0;
while read l;
do [[ $l =~ \$HESS ]] && f=1;
[[ f -eq 1 ]] && echo $l;
[[ $l =~ \$END ]] && f=0;
done < file
These other options are less readable to me, but seem to work nicely.
Freitag, 1. April 2011
Fails.
Donnerstag, 31. März 2011
Python 002: Version Introspection
One can obtain the version of the running Python instance by issueing
from sys import version_info
if version_info < (3,0):
print version_info
else:
print(version_info)
Vim 001: Editing Code
A part of the formatting also comes from introducing syntactic elements such as stars to group the code by content. In Vi this can be done in a simple way.
ESQ -> N -> a -> * -> ESQ -> RETURN
where N is the number of stars to be added, e.g. 40, or 78 to indicate the maximum width the code should take up.
Python 001: Module introspection
#!/usr/bin/env python3
import os.path
def getModuleName():
print("Current module:".ljust(30), os.path.abspath(__file__))
getModuleName()
Adding the above to the module should make it possible to get the location and name of the module in scope at runtime.
Mittwoch, 30. März 2011
PyMOL 003: Synchronizing Atoms in PyMOL
- Show both structures as lines
- Show the rank of the atoms.
- Follow the backbone ranks of both files until they loose synchronization.
- Adjust one to the other, from now on always adjust this one to the other. Only one of the files is changed.
PyMOL 002: Description of the Program
Also, PyMOL is not a modeling program (even though its modeling capabilities are already quite impressive). Instead, its a viewer and an analysing tool which is extensible. And this is the most important part about it I believe. It is extremely friendly to the user who wants to extend it by his or her own functionality. All that is required is some knowledge about Python. The PyMOL API is quite introspective, so every function documents itself and this is really makes the extension work convenient.
GAMESS 001: Files
Which file containes what, where to find coordinates from optimization jobs and how to efficiently transfer them to new input files.
There is a tool called DATAGAM which can fetch various $GROUP content from GAMESS files. It's not exactly clear to me how it works, but by issuing the following command, at least I was able to transfer the equilibrium geometry to a new file.
mzh @ ~/software/DATAGAM $ datagam -G 4-2-a-1-1-rm1--rm1.log
This is optimize run
Equilibrium geometry (with unique and all coordinates) will be saved to file OPTMZ
GAMESS 001: Files
Which file containes what, where to find coordinates from optimization jobs and how to efficiently transfer them to new input files.
Sonntag, 27. März 2011
Samstag, 26. März 2011
Schey 001: Bereichsintegrale
Schey macht sich viel Mühe bei der Behandlung von Bereichsintegralen. Wenn ich das soweit richtig verstehe, wird die Fläche S über die integriert wird, auf eine Ebene projeziert. Dann wird über diese Fläche integriert, wobei der Integrand mit dem Wert der Funktion auf der S Fläche multipliziert wird. Das Skalarprodukt aus dem Einheitsnormalvektor auf S mit dem Einheitsvektor in z-Richtung an der Stelle (x,y) liefert die Beziehung zwischen S und der Projektion R. Am Wochenende muss ich dann mal das Kapitel im Zachmann gründlich durchackern. Find das Thema schon ziemlich interessant.
Sonntag, 20. März 2011
PyMOL 001: Introduction to Indexing
One internal characteristics I would like to know more details about is, what the three labels Rank, ID and Index exactly mean and how they are related to the information contained in a PDB file. Work in progress.
Donnerstag, 17. März 2011
Shell 003: Data Handling
For a given system, the TS was first located in a subset of the system and then reinserted into the full system. Then the full system was reoptimized, but unfortunately the TS is no longer confirmed by a frequency calculation.
My guess is that the environment forces the TS into a local minimum. So the idea is not to let the TS readjust to the environment, but only the environment to the TS.
How to include this approach in the file naming?
On another aspect. The following is a situation which I encounter many times. I compute some structure which is part of a reaction. This structure now needs to be processed by different methods. Either as a starting point for a new method which leads to a new geometry, or a method which gives information about the quality of the structure as it is right now (its gradient, its hessian). For the files which arise from this structure, how do I indicate that they are in this "parent-child" relation?
Mittwoch, 16. März 2011
Hydrogens 001
This is probably something a lot of people require since its crucial for molecular modeling.
I have encountered several difficulties so far:
- Not all residues of a given type have the same pKa and therefore protonation state. How to tell the program so?
- Ionizable residues can be protonated at different positions (Nd or Ne in histidine).
It optimizes the histidine flips. It is a published algorithm, rather than the unsophisticated one that pymol uses, which is not really doing any optimization.
Dienstag, 15. März 2011
Anderson, J. M., Ex. 2.4
$f(x) = x-x^2$
on the interval
$[-\pi:\pi].$
What I'm asking myself is, what happens to the expansion coefficients
$c_m = \frac{1}{\pi}\left<\cos mx | f\right>$
$d_m = \frac{1}{\pi}\left<\sin mx | f\right>$
Another point of interest. The solutions in the book are
\[x-x^2= - \sum_{n { } even} \frac{4}{\pi n^3} \sin nx = \\ - \sum_{n even} \frac{4}{n^2} \cos + \sum_{n odd} \frac{4\pi-4}{\pi n^2}\cos n x + \left( \pi - \frac{2*\pi^2}{3}\right)" align="middle" alt="[latex]x-x^2= - \sum_{n { } even} \frac{4}{\pi n^3} \sin nx = - \sum_{n even} \frac{4}{n^2} \cos + \sum_{n odd} \frac{4\pi-4}{\pi n^2}\cos n x + \left( \pi - \frac{2*\pi^2}{3}\right)[/latex]" title="[latex]x-x^2= - \sum_{n { } even} \frac{4}{\pi n^3} \sin nx = - \sum_{n even} \frac{4}{n^2} \cos + \sum_{n odd} \frac{4\pi-4}{\pi n^2}\cos n x + \left( \pi - \frac{2*\pi^2}{3}\right)\]
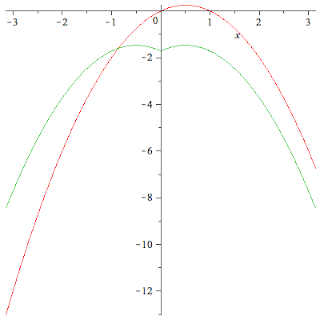
Offenbar war da ein Problem. c0 muss noch durch zwei geteilt werden. Ich dachte, dass sei bereits in der Lösung enthalten. Dadurch ergibt sich folgender Term
\[F := N \rightarrow -\sum_{n=1}^N \frac{4}{(2n)^2} \cdot \cos (2nx) + \\ { } \sum_{n=1}^N \frac{4\pi - 4}{\pi (2n-1)^2} \cos ((2n-1)x) + \frac{\pi - (2\pi^2/3)}{2}\]
Weiterhin bleibt zu vermerken, die Fourierreihe ist eine vollständige Menge orthonormaler Funktionen.
Shell 002: allsh
http://72.14.189.113/howto/shell/allsh/
Freitag, 11. März 2011
Donnerstag, 10. März 2011
Books I am working on
- Mathematics for Quantum Chemistry: Jay Martin Anderson
- div, grad, curl and all that: H. M. Schey
Best passage from Schey so far:
"Gauss' law is
\[\iint_S \textbf{E} \cdot \hat{\textbf{n}} \mbox{ dS} = \frac{q}{\epsilon_0}.\]
If you don't understand this equation, don't panic."
Isn't this just wonderful? All people coming from a German education background will know why I am posting this.
Shell 001: How to mv a big number of files efficiently
1scf-001.mop
1scf-002.mop
1scf-003.mop
1scf-004.mop
1scf-005.mop
1scf-006.mop
1scf-007.mop
1scf-008.mop
1scf-009.mop
1scf-010.mop
The question is, how can the files be changed such that both '1scf' and 'mop' are changed to 'mutant' and 'inp' on one command?
Various Checks
$E=mc^2?$
The good thing now is that
$\LaTeX$
is available in the blog.
Einstein most famous equation is \$E=mc^2\$.
Newtons derived the equaiton \\[s=ut+\sfrac{1}{2}at^2\\]
Einstein most famous equation is $E=mc^2$.
Newtons derived the equaiton \[s=ut+\sfrac{1}{2}at^2\]
Latex formating together with regular text inside 'pre' elements is still not working.
Check out this tutorial on inline mathematical notation.
http://watchmath.com/vlog/?p=438
Trying MathML from Maple:
Maple Latex Export
\[\mbox{}+1/2\,{\it cm}\,\cos \left( x \right)
+1/2\,{\frac {{\it cm}\, \left( \sin \left( x \righ t) \right) ^{2}}{\cos \left( x \right) -1}}
+1/2\,{\frac {{\it dm}\,\sin \left( x \right) \cos \ left( \left( N+1 \right) x \right) }{\cos \left( x \right) -1}}-1/2\,{\it dm}\,\sin \left( \le ft( N+1 \right) x \right)
\]
Motivation and Introduction to the Blog
From a quantum chemistry point of view, I most likely will be posting content related to GAMESS and MOPAC.
Finally, I plan to add solutions to various exercises I worked out from text books.

